Pseudomonas aeruginosa Counting Service
- Product Code: 127391
Pseudomonas aeruginosa Counting Service
description Service Overview
Pseudomonas aeruginosa Counting Procedure
Materials and Equipment
- Sample: e.g., water, environmental, or clinical sample.
- Sterile containers and pipettes
- Sterile dilution blanks (e.g., 0.1% peptone water or sterile saline)
- Membrane filtration apparatus
- 0.45 µm pore-size sterile membrane filters
- Selective Agar Plates: Typically, cetrimide agar (or another selective medium like Pseudomonas CN agar) is used.
- Incubator: Set at 35–37°C.
- Forceps: Sterile, for handling membrane filters.
- Disposable gloves and lab coat: For personal protection.
- Colony counter (manual or automated), if available
Procedure
-
Sample Preparation and Dilution
- a. Homogenize the Sample (if necessary):
If you are dealing with a complex matrix (e.g., food or soil), homogenize the sample in sterile saline or buffer. - b. Prepare Serial Dilutions:
To ensure countable colony numbers, prepare serial dilutions (e.g., 10^–1, 10^–2, 10^–3, etc.) using sterile dilution blanks. For water samples with expected low bacterial counts, you might filter the entire volume (e.g., 100 mL) directly without dilution.
- a. Homogenize the Sample (if necessary):
-
Membrane Filtration
- a. Assemble the Filtration Apparatus:
Set up the sterile membrane filtration system in a clean area (ideally in a biosafety cabinet if working with clinical or environmental pathogens). - b. Filter the Sample:
Filter a measured volume (e.g., 100 mL or an aliquot from your dilutions) through a 0.45 µm sterile membrane filter. If you are using dilutions, record the volume filtered for each dilution. - c. Transfer the Filter:
Using sterile forceps, carefully transfer the filter onto the surface of a cetrimide agar plate. Ensure the filter lies flat on the agar surface.
- a. Assemble the Filtration Apparatus:
-
Incubation
- a. Incubate the Plates:
Place the agar plates in an incubator set at 35–37°C. - b. Incubation Time:
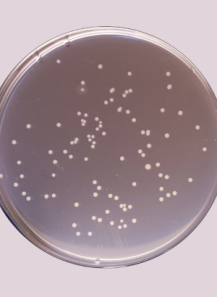
Incubate for 24 to 48 hours. P. aeruginosa typically produces characteristic colonies (often with a bluish-green pigment due to pyocyanin production) within this timeframe.
- a. Incubate the Plates:
-
Colony Identification and Counting
- a. Identify Colonies:
After incubation, inspect the plates for colonies that have the typical appearance of P. aeruginosa (rough, pigmented, sometimes with a fruity odor). Because cetrimide agar is selective, most colonies should be P. aeruginosa, but confirmatory tests (e.g., oxidase test, pigment production, Gram stain) may be warranted for regulatory purposes. - b. Count Colonies:
Choose the dilution plate that yields 30–300 colonies (if possible) for accurate counting. Count each colony (either manually or with an automated colony counter).
- a. Identify Colonies:
timeline Service Steps
| Step | Procedure | Expected Result |
|---|---|---|
| info Service steps will be provided upon request | ||
Cart
No products
Subtotal:
0.00
Total
0.00
THB